
DPhil Statistics
Machine Learning for Antibody Flexibility Prediction
Department of Statistics
University of Oxford
2021 - 2025
I am a DPhil student in the Oxford Protein Informatics Group at the Department of Statistics at Oxford University and an Industrial Fellow of the Royal Commission 1851. I am interested in developing developing deep learning methods for computational drug development. Currently, the production of new drugs requires tedious wet lab experiments that are associated with high costs. Computational methods have the potential to greatly facilitate the drug discovery process. Specifically, my research focuses on antibodies, a class of proteins of great interest to the pharmaceutical industry, and I am working on improving methods for predicting antibody structures and flexibility from sequence data.

Machine Learning for Antibody Flexibility Prediction
Department of Statistics
University of Oxford
2021 - 2025

NMR Study of Transmembrane Receptors
Department of Biochemisty
University of Oxford
2020 - 2022

Institute of Structural and Molecular Biology
University College London
2017 - 2020
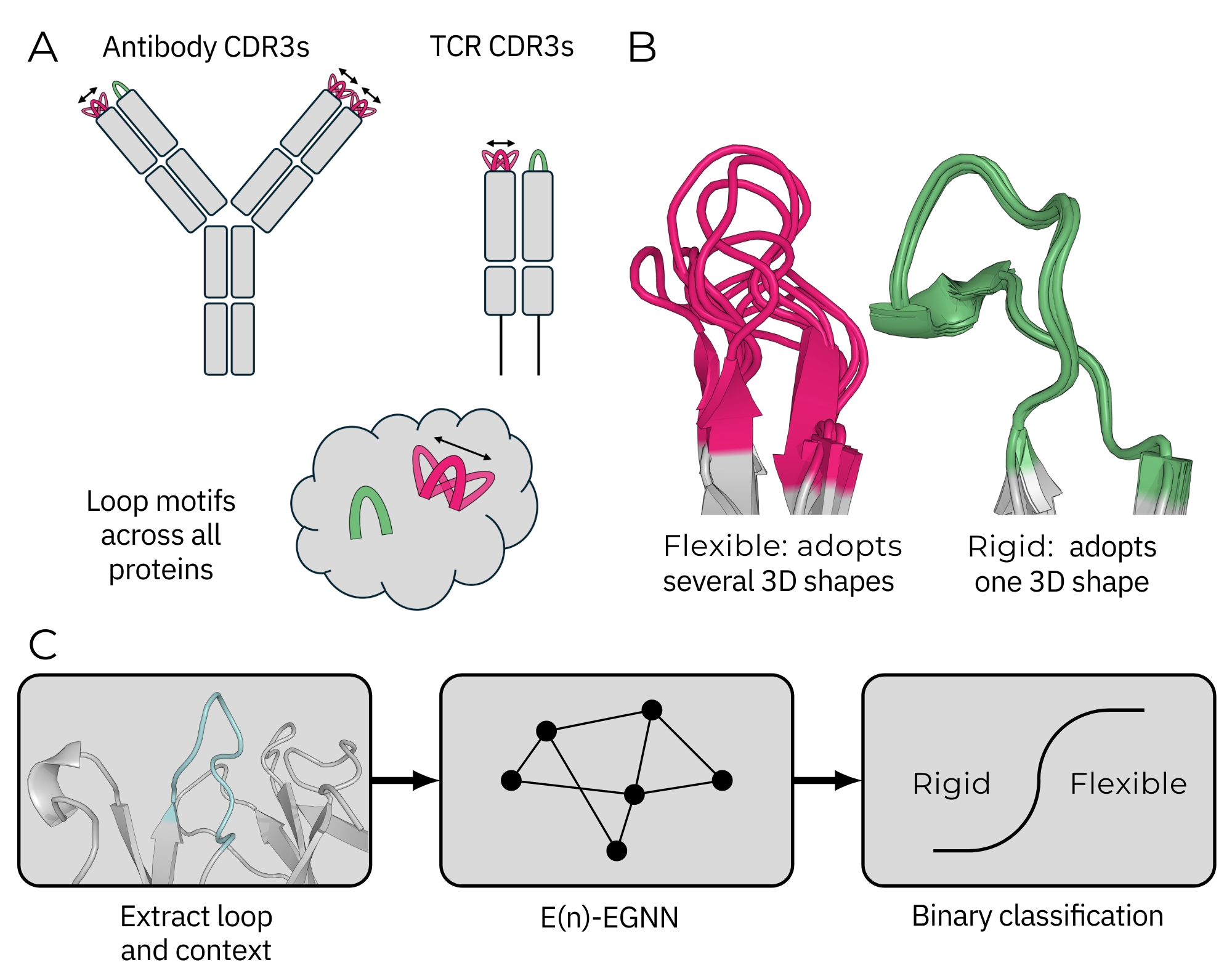
Cagiada M, Spoendlin FC*, Ifashe K and Dean CM. Nature Machine Intelligence, October 2025.
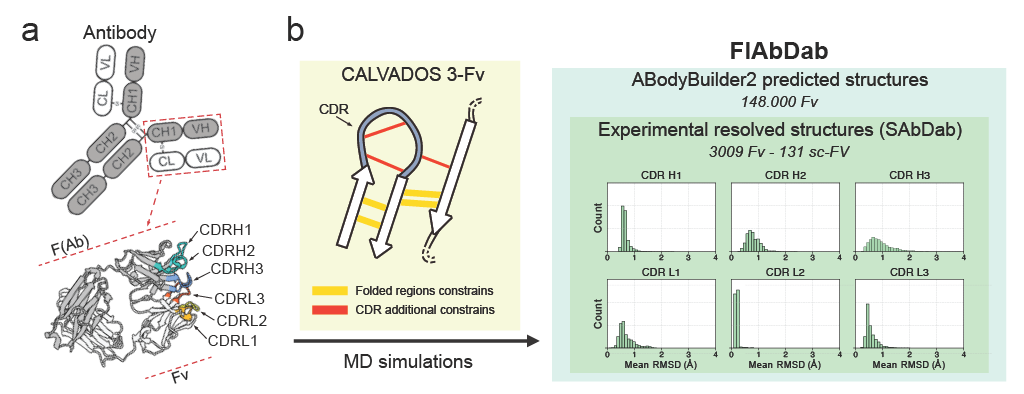
Spoendlin FC, Fernandez-Quintero ML, Raghavan SSR, Turner HL, Gharpure A, Loeffler JR, Wong WK, et al. Nature Machine Intelligence, October 2025.
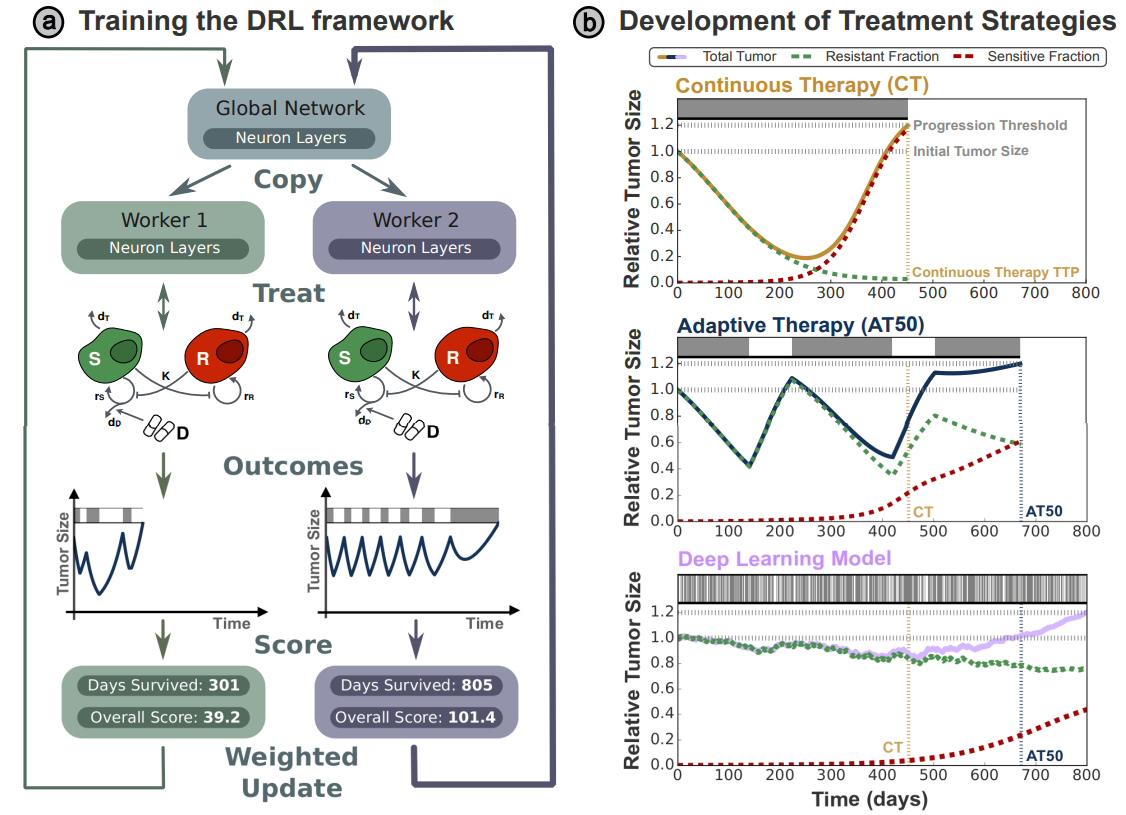
Gallagher K, Strobl MA, Park DS, Spoendlin FC, Gatenby RA, Maini PK, and Anderson AR. Cancer Research, April 2024.
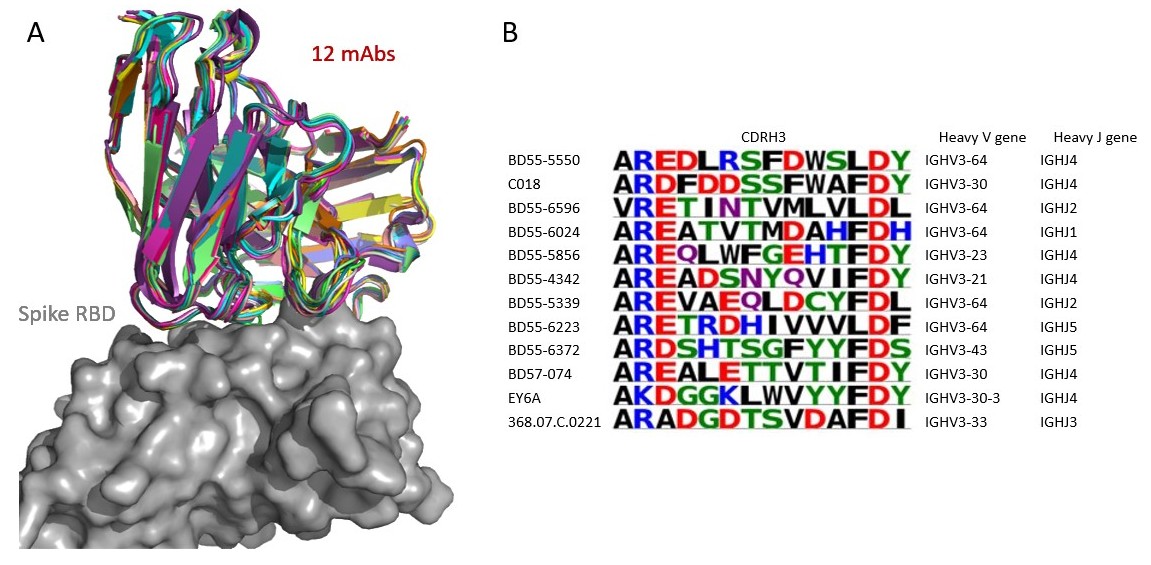
Spoendlin FC, Abanades B, Raybould MIJ, Wong KW, Georges G, and Deane CM. Frontiers in Molecular Biosciences, September 2023.
ANARCII: A Generalised Language Model for Antigen Receptor Numbering. Greenshields-Watson A, Agarwal P, Robinson SA, Williams BH, Gordon GL, Capel HL, Li Y, Spoendlin FC, et al. BioRxiv. April 2025.
Challenges and compromises: Predicting unbound antibody structures with deep learning. Greenshields-Watson A, Vavourakis O, Spoendlin FC, Cagiada M, and Deane CM. Current Opinion in Structural Biology, February 2025.
Assessing AF2's ability to predict structural ensembles of proteins. Riccabona JR*, Spoendlin FC*, Fischer MA, Loeffler JR, Quoika PK, Jenkins TP, Ferguson JA, et al. Structure, September 2024.
Sweeney C, Quast NP, Spoendlin FC and Teh YW. NeurIPS 2024 Workshop: Machine Learning in Structural Biology, December 2024.
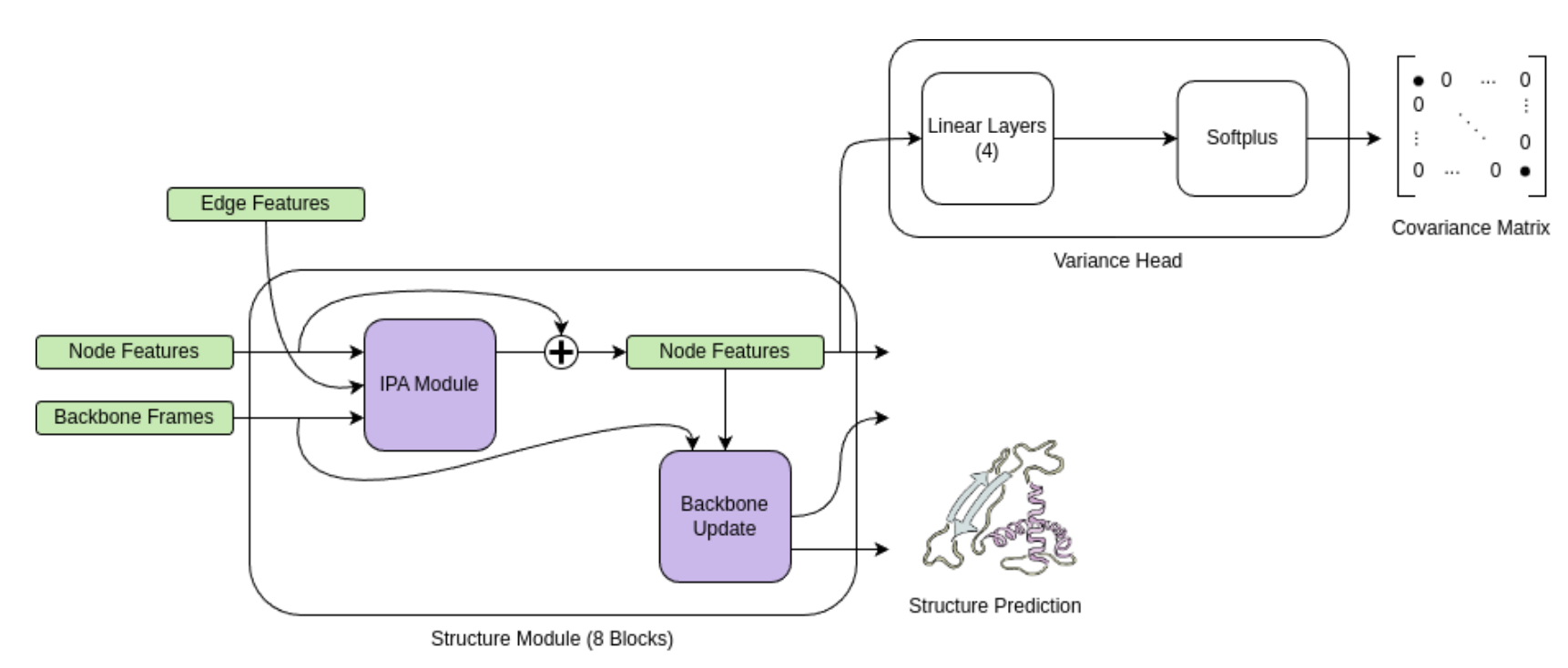
Spoendlin FC, Wong KW, Georges G, Bujotzek A and Deane CM. ICML 2024 Workshop: Machine Learning for Material and Life Sciences, June 2024.
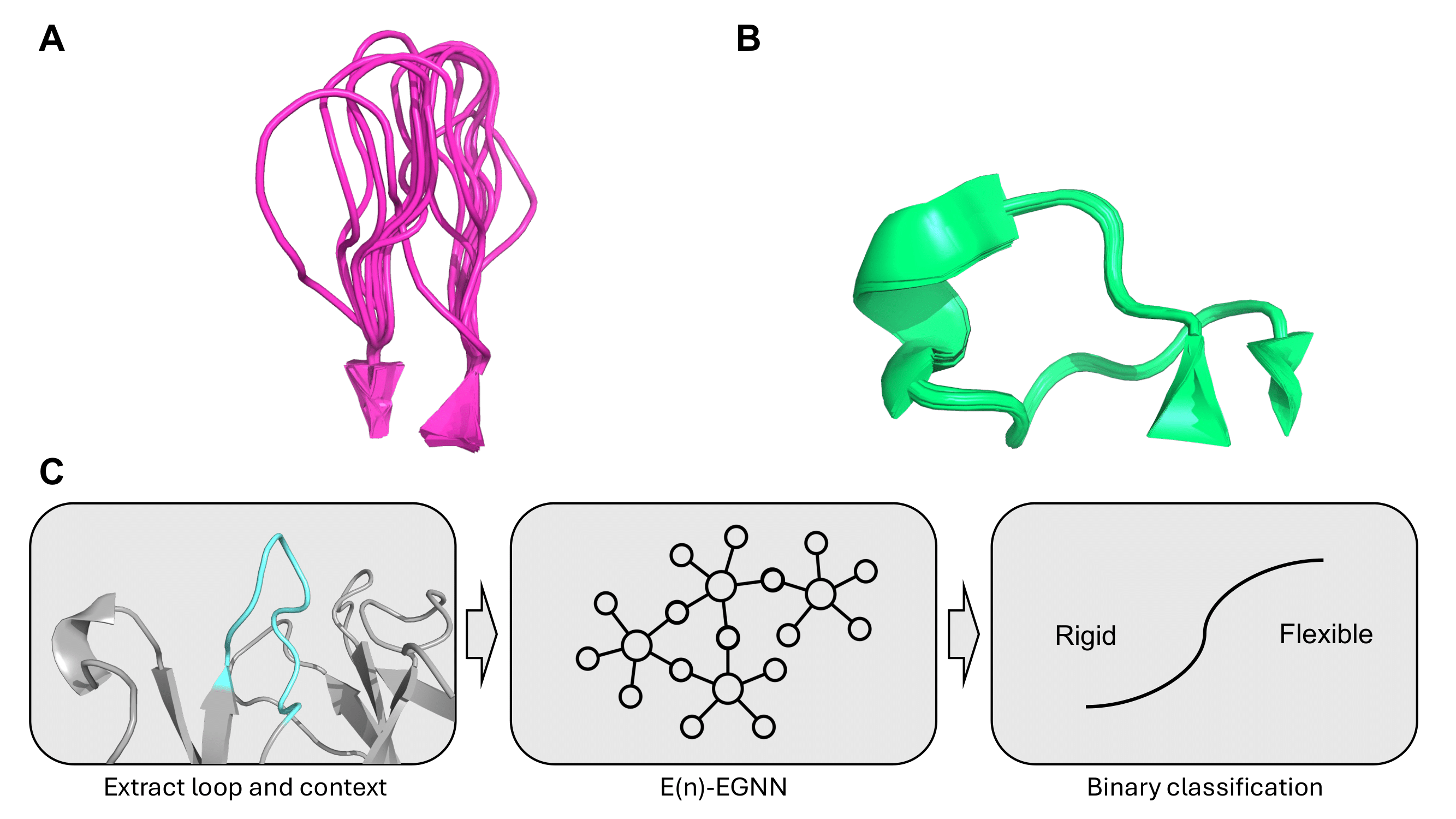
Structure-Based Epitope Profiling with the Structural Profiling of Antibodies to Cluster by Epitope 2 (SPACE2) Algorithm. Spoendlin FC and Deane CM. In Epitope Mapping Protocols. New York. NY: Springer US. 2025.
Investigating the dynamics of KDEL receptor signalling. Jan 2022.
Supervisors: Prof Jason Schnell & Prof Simon Newstead.